Movies
Research
Discovery of new drugs against African sleeping sickness, Chagas disease, and Leishmaniasis
A major aim of our research is the discovery of new drugs against trypanosomatids. Trypanosomatid parasite infections result in devastating diseases among the poor population in developing countries. Three of these diseases are caused by the protozoan parasites of the family trypanosomatidae Trypanosoma and Leishmania. African sleeping sickness, Chagas disease, and Leishmaniasis are caused by T. brucei, T. cruzi, and Leishmania respectively. Currently about twenty million people are infected. Owing to migration, T. cruzi infected population is also residing in the developed countries (100,000 in the EU and 300,000 in the US). Currently used drugs are toxic, lack efficacy against different stages of the disease and drug-resistant strains are emerging. No vaccine is available as these parasites evade host immune response by antigenic-variation or by hiding from the host immune defense.
Glycosomes are peroxisome-related organelles found only in Trypanosomatid parasites. The first seven enzymes of the glycolysis and also other essential metabolic pathways are compartmentalized inside the glycosomes. This compartmentation is essential since the parasite glycolytic enzymes lack feedback regulation. Their mislocalization to the cytosol leads to ATP depletion, accumulates glucose metabolites to toxic levels and kills the parasites. Glycosomal enzymes are recognized by the receptor PEX5 in the cytosol. The receptor-cargo complex docks at the glycosomal membrane by binding to PEX14. PEX5 and PEX14 together constitute a transient pore which transports the enzymes into the glycosome lumen. We report a way to selectively kill Trypanosoma by blocking glycosomal/peroxisomal import that depends on the PEX14-PEX5 protein-protein interaction. In cooperation with the Helmholtz-Institute Munich, we developed small molecules that efficiently disrupt the PEX14-PEX5 interaction. This results in mislocalization of glycosomal enzymes, causing metabolic catastrophe, and it kills the parasite. High-resolution x-ray structures and nuclear magnetic resonance data enabled the efficient design of inhibitors with trypanocidal activities comparable to approved medications. These results identify the glycosomal protein import machinery as an “Achilles’ heel” of the Trypanosoma suitable for the development of new therapies against trypanosomiases and provide the structural basis for their development. As a proof-of principle, we could show that small molecule inhibitors of receptor docking to the peroxisomal membrane disrupt glycosomal protein import and kill Trypanosoma parasites (Dawidowski et al.,2017).
Protein import into Peroxisomes
Protein sorting plays a fundamental role in the biogenesis and maintainance of peroxisomes. Proteins resident in the peroxisomal matrix are encoded by nuclear genes, synthezised on free ribosomes, and imported post-translationally into the organelle. The transport requires both ATP and cytosolic factors and recent evidence suggests that peroxisomal proteins can be imported in a folded state. Genetic and biochemical evidence indicates that two pathways are involved in the sorting process of peroxisomal matrix proteins, corresponding to the existence of two distinct peroxisomal targeting signals, the C-terminal PTS1 and N-terminal PTS2. Recognition of PTS1- and PTS2-targeting signals is performed in the cytosol by the PTS-specific import receptors, Pex5p and Pex7p, respectively. The widely accepted ‘extended shuttle’ model of peroxisomal protein import suggests that the import receptors Pex5p and Pex7p bind cargo proteins in the cytosol, dock to specific proteins at the periphery of the peroxisomal membrane, subsequently enter the peroxisome, release their cargo in the lumen of the peroxisome, and shuttle back to the cytoplasm. We have identified and characterized components of a docking complex for both signal recognition factors (Pex13p, Pex14p and Pex17p, see ) indicating that the two pathways are not independent but overlap at the level of the peroxisomal membrane. A main line of our research is the functional characterization and structural analysis of components of the peroxisomal protein import machinery. Components of the import machinery will be crystallized and subjected to structural analysis. In vivo and in vitro are used to study the functional interactions of the proteins involved in the recognition and transport of peroxisomal proteins. We also apply new methods to identify new components of the import machinery and we develope an in vitro system to study the mechanism of the translocation process.
Biogenesis of the Peroxisomal Membrane
We recently discovered that the post-translational targeting of peroxisomal matrix and membrane proteins from the cytosol to peroxisomes is performed by distinct pathways. The identification and characterization of components of the pathway for the post-translational targeting of peroxiosmal membrane proteins is a line of research in our laboratory. However, recent evidence suggests the existence of a new protein transport route to peroxisomes via a preperoxismal compartment which might be the endoplamic reticulum. A subset of peroxisomal membrane proteins might be targeted first to the endoplasmic reticulum and subsequently to peroxisomes by a vesicle-mediated transport (see). Following the intracellular routing of selected membrane proteins and by functional analysis of suspected components of this pathway, we indend to contribute to the elucidation of this vesicle-mediated transport of peroxisomal membrane proteins.
Peroxisomal Metabolism and Metabolite Transport
We established a reverse genetic approach to identify yeast peroxisomal proteins which combines traditional cell fractionation with modern computer techniques. Peroxisomal matrix and membrane proteins were isolated by subcellular fractionation and extraction techniques and isolated proteins were subjected to microsquencing and mass spec analysis. As the yeast genome is sequence, the generated sequence or mass spec data usually were sufficient to identify the corresponding protein by computer analysis. So far, we identified more than 20 new peroxisomal matrix and membrane proteins, comprising new peroxisomal enzymes as well as metabolite transporters. The functional analysis of these proteins will improve our knowledge on peroxisomal metabolism and metabolite transport across the peroxisomal membran.
Proliferation of Peroxisomes
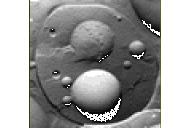
Peroxisomes are supposed to multiply by growth and division. In line with this assumption, we identified Pex11p, deficiency of which results in the generation of giant peroxisomes in yeast (see). This phenotype indicated that Pex11p is required for the proliferation of peroxisomes. Pex11p is the first and only component identified which is required for this cellular process. In an attempt to identify other proteins of this type, we currently apply a genome wide screen for peroxisomal mutants with abberant peroxisome morphology.